In the hunt for improved antibiotics, it is important to realize that natural products have been, without question, the most prolific source of all medicines. With the advent of massively parallel DNA sequencing, it has become apparent that our knowledge of natural product structure and function is astonishingly incomplete. Exploration of uncharted natural product chemical space will undoubtedly lead to improved, and entirely new, medicines. Thus, our group focuses on elucidating the biosynthesis, structure, and function of natural products. Our primary focus has been on thiazole/oxazole-containing peptides. Characterized examples have activities ranging from antibacterials to virulence-promoting toxins. Therefore, the study of these peptidic natural products allows us to not only better understand bacterial virulence (where pharmacological intervention would constitute a pathogen-specific approach to bacterial infection) but also explore unique chemical architectures, ideally positioning us to introduce new structural classes of antibiotics. Our recent successes and focal areas are highlighted below by research project.
lasso peptides
We bioinformatically mapped lasso peptide clusters and structures on a large scale and are using this information to study these antibacterial peptides
recent publications
- Kretsch, A.M.; Gadgil, M.G.; DiCaprio, A.J.; Barrett, S.E.; Kille, B.L.; Si, Y.; Zhu, L.; Mitchell, D.A. "Peptidase activation by a leader peptide-Bound RiPP recognition element." Biochemistry, 62 956-967 (2023). doi: 10.1021/acs.biochem.2c00700
- Si, Y.; Kretsch, A.M.; Daigh, L.M.; Burk, M.J.; Mitchell, D.A. "Cell-Free biosynthesis to evaluate lasso peptide formation and enzyme–substrate tolerance." J. Am. Chem. Soc., 143: 5917–5927 (2021). doi:10.1021/jacs.1c01452
- Harris, L.A.; Saint-Vincent, P.M.B.; Guo, X.R.; Hudson, G.A.; DiCaprio, A.J.; Zhu, L.; Mitchell, D.A. "Reactivity-Based Screening for Citrulline-Containing Natural Products Reveals a Family of Bacterial Peptidyl Arginine Deiminases." ACS Chem. Biol., 15: 3167–3175 (2020). doi:10.1021/acschembio.0c00685
Lasso peptides have a variety of characterized bioactivity and hold great promise as a therapeutic scaffold due to their exemplary stability to proteases and extreme environments. To facilitate their discovery from natural sources, we developed Rapid ORF Description and Evaluation Online (RODEO) which automates the time-consuming process of annotating biosynthetic gene clusters and predicting likely lasso peptide precursors. Using RODEO, we have predicted thousands of lasso peptide gene clusters present in GenBank, the most comprehensive list to date. With this set of all lasso clusters and precursor peptides, we are poised to expand what is known regarding this expansive natural product family. We aim to discover and characterize lasso peptides with novel post-translational modifications, structures, and bioactivity.
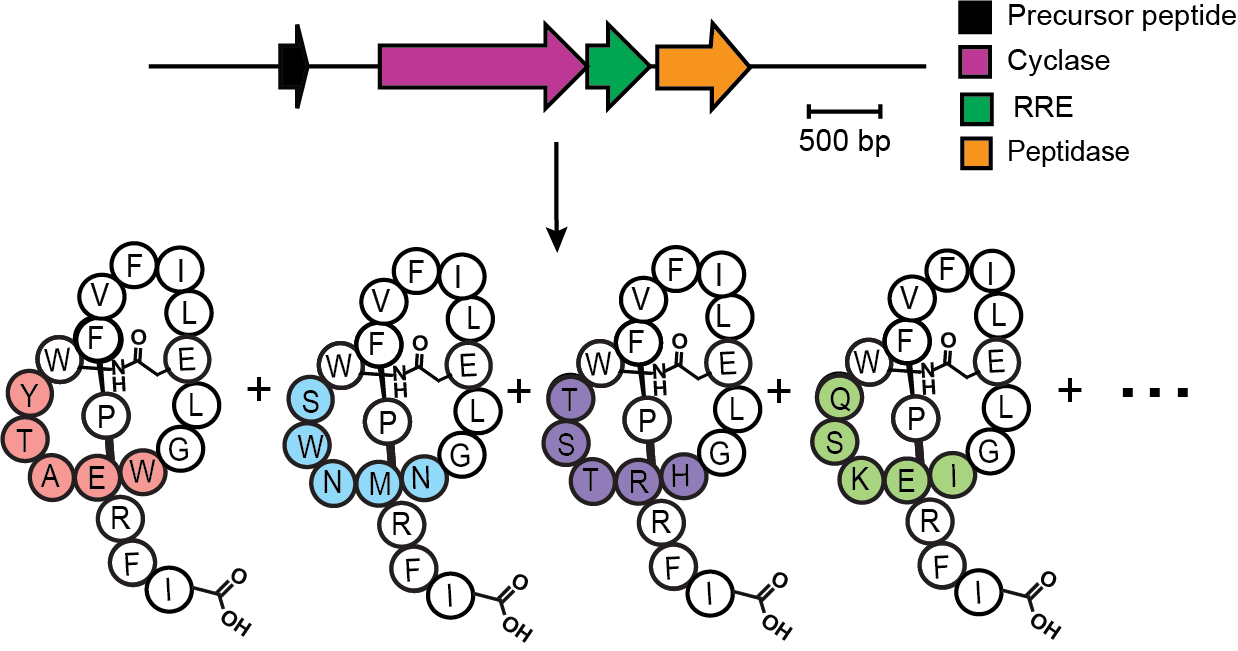
Despite our ability to accurately predict lasso peptide gene clusters, we understand little about the enzymology of the core biosynthetic enzymes. In particular, many questions remain about how the cyclase facilitates lasso peptide folding and ties the lasso knot. In 2019, RODEO enabled the discovery and characterization of the lasso peptide Fusilassin. This thermophilic derived lasso peptide system is only the second to date compatible with in vitro experiments, opening the doors for in-depth biochemical characterization of its enigmatic biosynthetic enzymes. Using cell-free technology, we were able to show that Fusilassin’s biosynthetic enzymes are extremely promiscuous, tolerating approximately 2 million diverse ring sequences. Combining the vast natural diversity of lasso peptides with their cyclase’s promiscuous substrate tolerance will enable the creation of customized, unnatural lasso peptides with interesting biological activity.